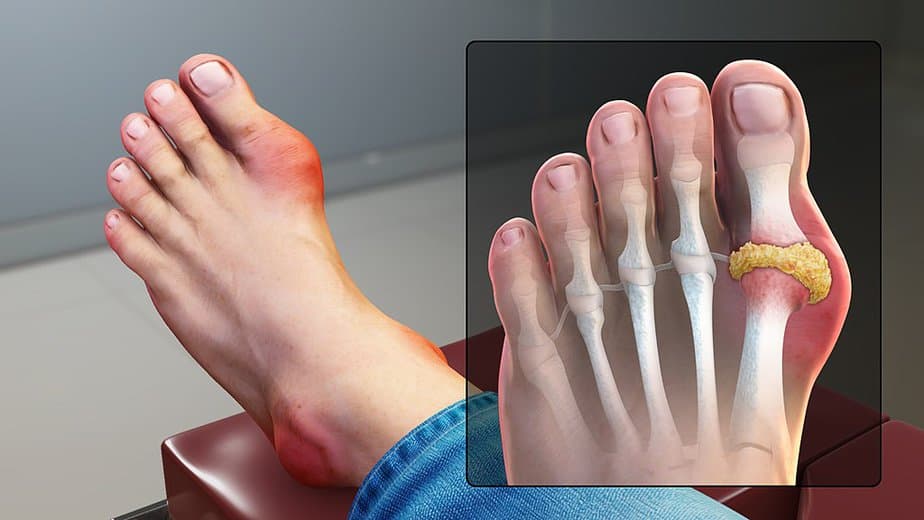
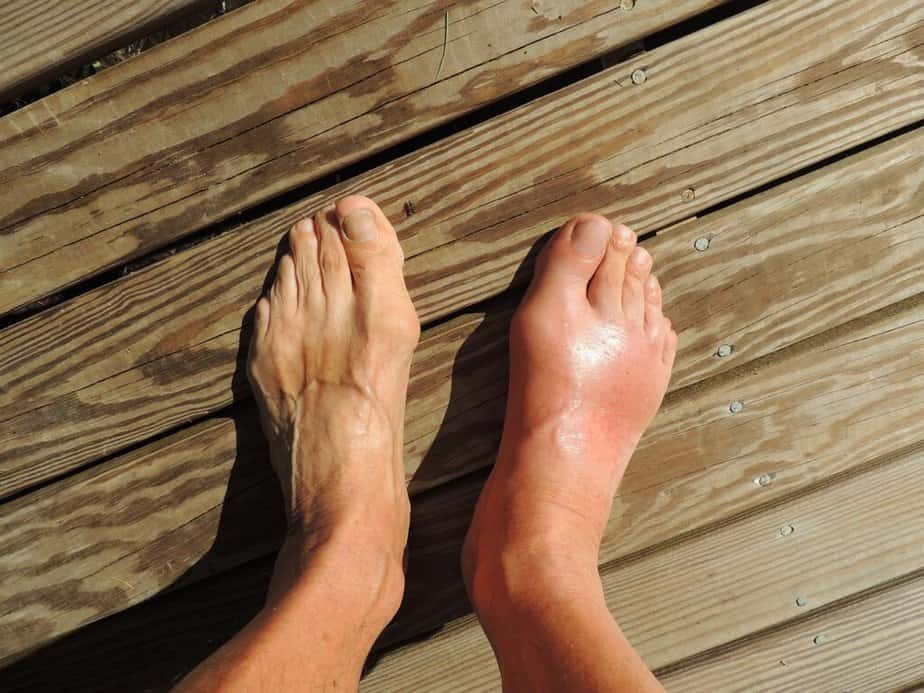
Gout, a common form of inflammatory arthritis, is characterized by intense joint pain, redness, tenderness, and swelling. Typically, gout occurs at the base of the big toe and may cause acute bouts of pain, known as flares, which usually subside after one to two weeks. Often, those experiencing recurrent episodes seek medical help.
Uric acid accumulation, leading to the formation of sharp urate crystals in the joints and tendons, causes gout. The condition is more common among individuals with a diet rich in animal products and alcohol and those genetically predisposed to it. To diagnose gout, a doctor will usually perform a blood test to measure uric acid levels. Then, they focus on treatments that reduce symptoms and lifestyle changes to lessen the likelihood of recurrence.
Numerous genome-wide association studies have explored the impact of genetic factors on gout susceptibility. Such studies have revealed genes involved in urate transportation, release, or reabsorption, sugar metabolism, and the transporting of other small molecules. ABCG2, a commonly implicated gene, codes for a protein that facilitates urate transport into the gut, aiding its elimination from the body. However, scientists believe there is more to discover about the genes influencing uric acid levels in the body.
The Study
A recent article published in Rheumatology points to URAT1 (SLC22A12), a gene involved in urate reabsorption, as significantly contributing to high urate levels. However, our understanding of how URAT1 variants affect gout risk remains limited. The authors discovered that both common and rare variants in the URAT1 gene offer significant protection against gout, illustrating promising prospects for personalized medicine.
The authors enrolled 480 Japanese men diagnosed with primary gout, and a control group of 480 healthy adults without hyperuricemia or a history of gout. Using targeted exome sequencing, the team identified variants linked with gout predisposition or prediction against gout.
In total, the authors identified one common and ten rare variants in the URAT1 gene. To understand how these variants affect uric acid levels and gout, they first evaluated their impact on how well the protein encoded by the URAT1 gene works.
Then, they investigated how the variants affected uric acid levels in blood samples and whether any of the variants were associated with an anti-gout effect.
Results
Of the eight variants used in the functional analysis, the authors discovered multiple variants that prevented URAT1 from functioning or significantly reduced its activity.
When looking at the rare variants only, the authors found that 2.5% of participants without gout carried one of these rare variants. Furthermore, the serum urate concentrations in those having a variant were significantly lower than those without the variants.
Since the effect of being a carrier is less uric acid buildup, the variants may also provide gout protection. This result is what the authors saw in their cohort. That is, having one of these rare variants drastically reduced the risk the individual would be clinically diagnosed with gout, even in the presence of the gout gene ABCG2.
This study is the first to reveal multiple rare anti-gout variants of URAT1. The results indicated that carrying these variants offer protection against gout and offer new perspectives in gout research and potential treatment avenues.
Explore your Genome!
Genome Browser
Did you know you can use the Nebula Genome Browser (available with Deep and Ultra Deep WGS) to check whether you have protective variants?
- Go to the Genome Browser. In the top left corner of the genome browser, you can find a search bar.
- The authors identified multiple independent variants in SLC22A12 that contribute to protection. The variants and their locations in the genome are listed in the table below.
- Copy-paste one of these locations into the search bar and press enter.
- The genome browser will now zoom in on these locations.
- Activate the “Center Line” in the bar at the top to better see the location that you are looking at.
- You should see stacked, gray stripes. Those are your personal DNA sequencing reads that are aligned to a reference genome sequence (colored letters above). If your DNA sequence matches the reference, then the stripes are gray. If the sequence is somehow different from the reference, then you will see various letters and symbols in different colors.
- The four variants, their locations, and protective alleles are shown in the table below:
| rsID | Location (GRCh38) | Reference allele | Protective allele |
| rs121907892 | 11:64593747 | G | A |
| rs121907896 | 11:64591825 | G | A |
| rs1286450383 | 11:64593438 | C | A |
| rs2039330411 | 11:64598574 | C | T |
| rs2039388764 | 11:64599742 | C | G |
| rs765990518 | 11:64599750 | A | T |
| rs121907895 | 11:64599858 | T | G |
| NA | 11:64601525 | T | Deletion |
Gene Analysis Tool
You can also use the Nebula Gene Analysis Tool (available with Deep and Ultra Deep WGS) to see whether it flags any additional variants.
This tool empowers you to examine any gene in your genome and identify important genetic variants and mutations.
- When you click on the “Get Started” button your VCF file will be loaded into the Gene Analysis tool in a new tab.
- Type “SLC22A12” into the search bar at the top.
- The Gene Analysis tool will extract genetic variants in the SLC22A12 gene from your VCF file and display them to you using symbols that have different colors. The colors denote the potential importance of variants. The Gene Analysis tool determines this by referencing the ClinVar database and other resources.
Any red and orange variants could be important. Click on them to check if any of them are truncating variants like the ones described in the study described above.
Citation
Toyoda Y, Kawamura Y, Nakayama A, Nakaoka H, Higashino T, Shimizu S, Ooyama H, Morimoto K, Uchida N, Shigesawa R, Takeuchi K, Inoue I, Ichida K, Suzuki H, Shinomiya N, Takada T, Matsuo H. Substantial anti-gout effect conferred by common and rare dysfunctional variants of URAT1/SLC22A12. Rheumatology (Oxford). 2021 Nov 3;60(11):5224-5232. doi: 10.1093/rheumatology/keab327. PMID: 33821957; PMCID: PMC8566256.
