Introduction to SARS
The severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2003 in China. An epidemic followed, spreading across 26 countries and infecting over 8000 people before the virus was contained. The typical symptoms of SARS are similar to the flu, including fever, headache, and overall feelings of discomfort. Severe cases of the illness were common, resulting in a mortality rate of nearly 10%. Because of the severity of the symptoms, it was possible to identify those infected with SARS-CoV and isolate them. Public health measures like social distancing were successful at containing the outbreak.
SARS and the COVID-19 pandemic
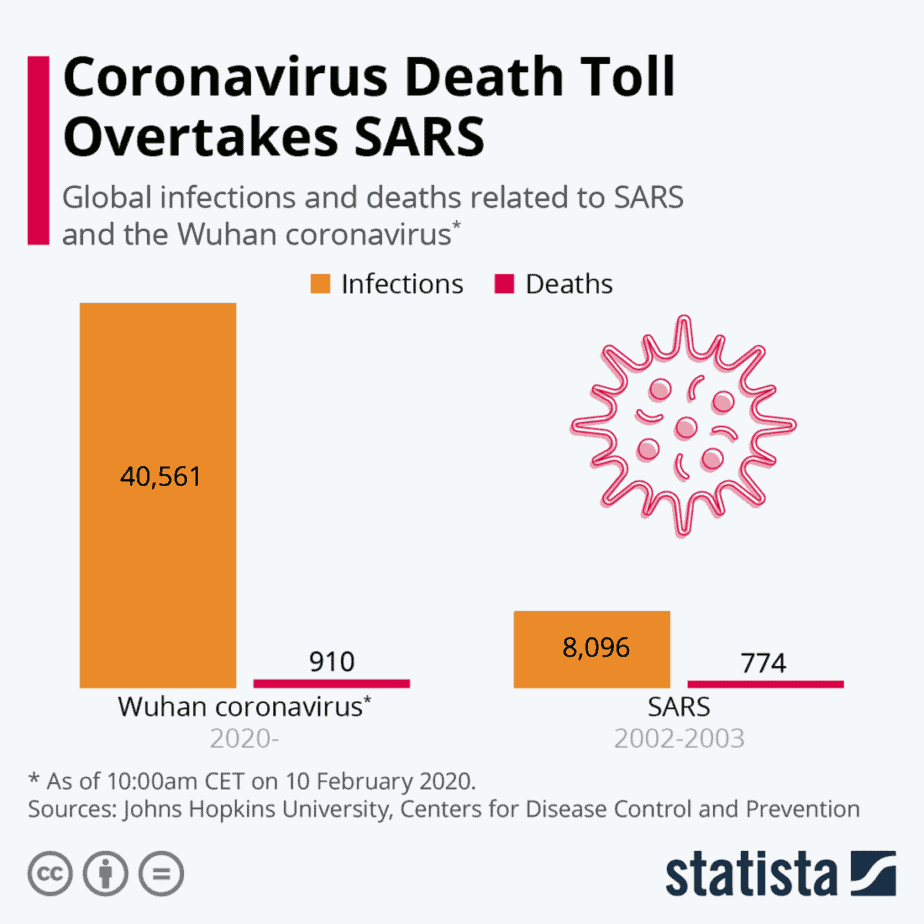
The situation is different with today’s COVID-19 pandemic. This novel coronavirus (SARS-CoV-2) can infect people who then remain largely asymptomatic. This makes it much more difficult to identify and contain those infected who continue spreading the disease. Although the mortality rate of COVID-19 is estimated to be 5 to 10 times lower than SARS, the total number of deaths has already surpassed the SARS outbreak due to failure to contain the pandemic.
However, SARS-CoV and SARS-CoV-2 also share many similarities. Both viruses originated in bats and share 80% of their genetic code. Furthermore, both viruses infect the respiratory tract using the same mechanism, and they cause similar symptoms.
While our understanding of COVID-19 is still lacking, much research on SARS has been produced in the years after 2003. Researchers have identified genetic variants that are associated with susceptibility to infection with SARS-CoV. It remains an open question whether the same genetic variants are also associated with susceptibility to SARS-CoV-2 infection given the similarities between the two coronaviruses.
To inform our users about past research on coronaviruses, we added 3 studies on SARS coronavirus infection to the Nebula Research Library. These studies give us some understanding of the links between human genetics and coronavirus infection.
Nebula Library entries on SARS
SARS coronavirus infection (Hamano, 2005)
This study explored genetic data from Vietnamese individuals and identified a genetic variant in the OAS-1 gene that is associated with susceptibility to infection with the SARS coronavirus.
SARS coronavirus infection (Ching, 2010)
This study used genetic data from Chinese individuals to demonstrate that a genetic variant is linked to a decreased susceptibility to SARS coronavirus infection. This variant is found near the MxA gene, which normally plays a role in inhibiting the replication of the SARS coronaviruses.
SARS coronavirus infection (Tu, 2015)
This study examined the genetic data of Chinese SARS patients to identify variants in 2 genes that lead to an increased risk of infection. One variant is in the CCL2 gene, which plays a role in attracting macrophages to an infection site. The other variant is in the MBL gene, which helps the immune system recognize molecular patterns that are commonly found on the surface of many viruses and bacteria.
New COVID-19 research
Before long, studies on SARS-CoV-2 susceptibility and the role of genetics will be published. Here at Nebula, we are monitoring the research on COVID-19 and will provide our users with timely updates. You can stay updated with the latest research discoveries by purchasing our whole-genome sequencing or uploading your existing 23andMe or AncestryDNA data.
For updates on the genetics of COVID-19 visit: https://nebula.org/blog/category/science/
Interested in COVID-19 travel tests? Kurix Health offers special testing meeting requirements for departing and arriving in the UK.
You can also learn more about coronavirus tests in this blog post.