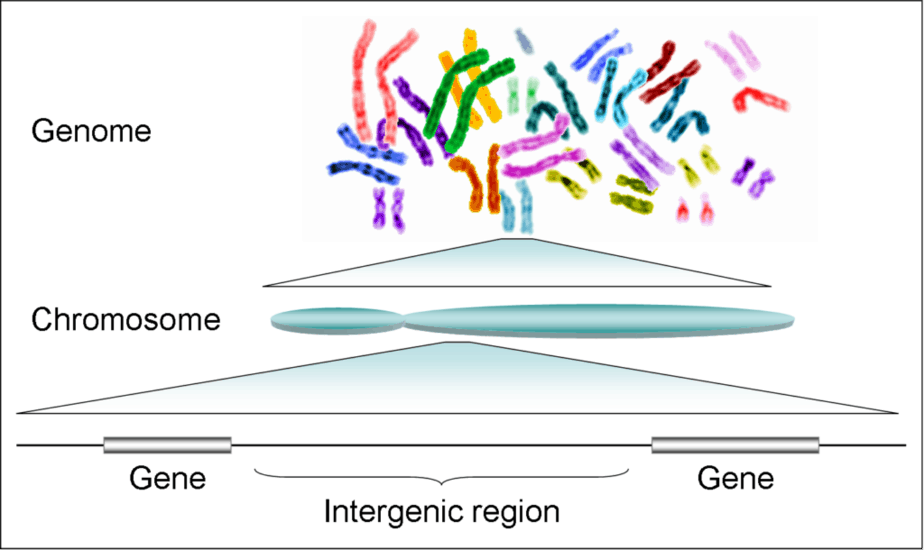
What’s a genome?
The genome is the whole material that carries the hereditary information of a cell or a virus particle. It exists as chromosomes, DNA, or RNA in the case of RNA viruses. In an abstract sense, this also includes the entirety of the hereditary information of an individual.
The term “genome” was coined by Hans Winkler in 1920. Genome research usually investigates structural variation and the interactions between genes.
Edited by Christina Swords, Ph.D.
Basics
The information required for the inheritance of traits is contained in DNA. More specifically, it lies in the sequence of the DNA bases adenine (A), guanine (G), cytosine (C) and thymine (T). Ribonucleic acids use the base uracil (U) instead of thymine. According to the rule of the genetic code, three consecutive bases each mean one amino acid.
A distinction is made between coding and non-coding sections of DNA. Proteins are formed from amino acids in the course of gene expression according to the base sequence of the coding sections. However, non-coding regions can also have important functions, for example in gene regulation. There are also pseudogenes: genes that have become functionless due to mutations and can no longer be read by the organism.
In addition to the DNA in the nucleus, further genetic material can be found in other parts of the cell. In eukaryotes, small genome sequences of their own are found in the mitochondria (mitogenome, also chondriome). In algae and land plants, they are almost always found in the chloroplasts and other plastids (plastome). Prokaryotes (bacteria and archaea) often contain additional, relatively short, self-contained DNA molecules called plasmids.
Organization of genomes
Eukaryotes
In eukaryotes, the nuclear genome (karyoma) consists of several to numerous strand-like chromosomes. The nuclear DNA is also called nuclear DNA (nDNA). The number of chromosomes is species-specific and can vary from two (in the horseworm) to several hundred (in some ferns).
The number of chromosomes also changes when the nuclear phase changes (meiosis and karyogamy). Eukaryotic genomes also have a high proportion of non-coding DNA and the intron-exon structure within its genes.
Prokaryotes
In prokaryotes, the DNA is present as a long, self-contained molecule. In addition, shorter, likewise self-contained DNA molecules, so-called plasmids, can be present in variable numbers. These can be amplified independently of the main DNA and passed on to other prokaryotic cells (conjugation), even across species boundaries. As a rule, they contain only a few genes, which for example mediate resistance to antibiotics.
Prokaryotic genomes are generally much smaller than eukaryotic genomes. They contain relatively small non-coding parts (5-20 %) and also only few or no introns.
Organelles
The genomes of the mitochondria, plastids, and other types of hydrogenosomes are organised like prokaryotes’. According to the endosymbiotic theory, those organelles are thought to exist previously as free-living prokaryotes. These ‘mitogenomes’ and ‘plastomes’ also contain a small part of the genes required for their own function. This is why these organelles are called “semi-autonomous”.
Viruses
Viral genomes are very small, since they contain only a few proteins. The genetic information is highly condensed because different genes overlap. Some sections can also function as genes in different sequence’s reading directions at the same time. The viral genome (also known as virom) can
- consist of DNA or RNA,
- this can be single or double stranded,
- be linear or circularly closed, and
- are segmented into several parts (multipartite) or unsegmented (monopartite)
Retroviruses are a special feature. Their RNA genome can be “translated” into DNA by reverse transcription and integrated into the host genome. The properties of the genomes of the viruses are important criteria for their classification.
Some viruses and especially virophages (viruses that attack other viruses) have mobile genetic elements (transposons, transpovirons, polintons). In general, their totality is also called a mobiloma.
Viroids
The genomic RNA of viroids is short. It spans between 241 and 401 nucleotides and contains many complementary regions that form double-stranded secondary structures. Viroids have no additional envelope and are 80 to 100 times smaller than the smallest viruses. They reproduce within living cells of higher plants.
Genome sizes
Genome size is the amount of DNA present in a genome. In eukaryotes, this information usually refers to the haploid set of chromosomes, which is also called the C-value. Either the number of base pairs present (bp) or the mass of DNA in the unit pg (picogram) is given.
One pg of double-stranded DNA consists of about 0.978-109 bp, i.e. almost one billion base pairs. Different organisms have different picograms to base pairs ratio nonetheless and the two units are not always easy to compare.
With the ease of genome sequencing in the last decade, measuring genome size in base pairs is more common. To sum 1,000 base pairs, the term “kilobase pairs” (kbp or kb) is usually used. For one million base pairs, we use “megabase pairs” (Mbp or Mb).
In 1972, the Ethiopian Lungfish (Protopterus aethiopicus) is often cited as a vertebrate with the largest genome at about 133 pg. In 2014, the record was broken by common locust (Locusta migratoria) with 6.3 Gbp. In 2018, 32 billion base pairs were detected from the Mexican axolotl (Ambystoma mexicanum).

The leaf flea endosymbiont Carsonella ruddii has the smallest bacterial genome quantified in 2006. Its circular DNA molecule contains only about 160,000 base pairs, which contain all the information it needs to live.
The DNA of a single human cell is about 1.80 m long when strung together. Theoretically, a base pair has an information content of 2 bits because it can assume 4 states (A/T/G/C). With about 3.27 billion base pairs, its maximum information content would be 6.54 billion bits or 780 MiB.
The actual information content is presumably lower as large parts of the DNA contain non-coding sequences with partial regulatory functions. According to the results of the Human Genome Project in 2003, 99.99% of the human genome contains genes.
There is a lack of correlation between the genome size and the complexity of the organism, or the “C-value paradox”. For example, caudates have larger genomes than reptiles, birds and mammals. Lungfish and cartilaginous fish have larger genomes than true bony fish. Within flowering plants or protozoa, however, the genome size varies considerably.
The largest amount of DNA is found in simple eukaryotes such as amoebae and the primeval ferns. These organisms have about one trillion base pairs. Their genome also contains individual genes as thousandfold copies and long non-protein-coding sections.
