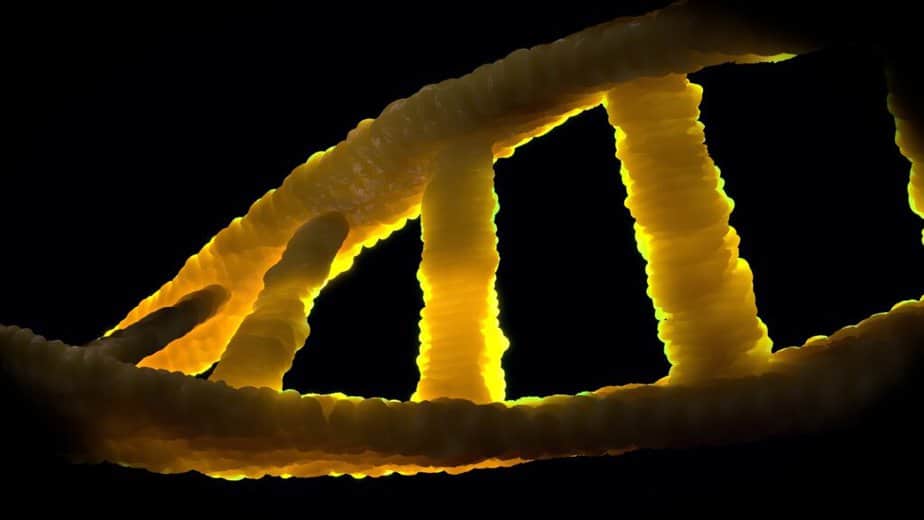
Intro to DNA
Deoxyribonucleic acid is a nucleic acid that is a polynucleotide composed of a chain of many nucleotides. The biomolecule located in the chromosomes is the carrier of genetic information in all living organisms and in many viruses (DNA viruses, pararetroviruses), i.e. the material basis of the genes.
In the normal state, DNA is constructed in the form of a double helix. Its building blocks are four different nucleotides, each consisting of a phosphate residue, the sugar deoxyribose and one of four organic bases (adenine, thymine, guanine and cytosine, often abbreviated as A, T, G and C).
The genes in the DNA contain the information for the production of ribonucleic acids. In protein-coding genes, this is an important RNA group, mRNA. It in turn contains the information for the construction of proteins, which are necessary for the biological development of a living being and the metabolism in the cell. The sequence of bases here determines the sequence of amino acids of the respective protein: The genetic code encodes a specific amino acid with three adjacent bases each.
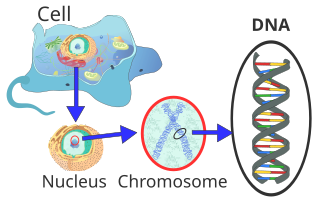
In the cells of eukaryotes, which also include plants, animals and fungi, the majority of the DNA in the cell nucleus (Latin nucleus, therefore nuclear DNA or nDNA) is organized as chromosomes. A small part is located in the mitochondria, the “power stations” of the cells, and is accordingly called mitochondrial DNA (mtDNA). Plants and algae also have DNA in photosynthetic organelles, the chloroplasts or plastids (cpDNA). In bacteria and archaeae – the prokaryotes that have no cell nucleus – the DNA is located in the cytoplasm. Some viruses, so-called RNA viruses, store their genetic information in RNA instead of DNA.
Edited by Christina Swords, Ph.D.
Structure and Organization of DNA
Building blocks
Deoxyribonucleic acid is a long chain molecule (polymer) made up of many building blocks called deoxyribonucleotides or nucleotides for short. Each nucleotide has three components: Phosphoric acid or phosphate, the sugar deoxyribose and a heterocyclic nucleobase or base. The deoxyribose and phosphoric acid subunits are the same for each nucleotide. They form the backbone of the molecule. Units of base and sugar (without phosphate) are called nucleosides.
The phosphate residues are hydrophilic due to their negative charge; they give DNA in aqueous solution an overall negative charge. Since this negatively charged DNA dissolved in water cannot give off any further protons, it is strictly speaking not (no longer) an acid. The term deoxyribonucleic acid refers to an uncharged state in which protons are attached to the phosphate residues.
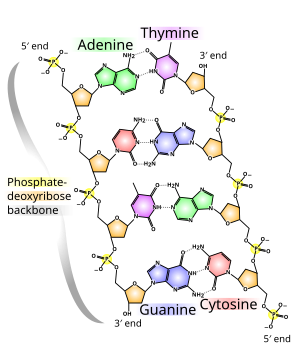
The base can be a purine, namely adenine (A) or guanine (G), or a pyrimidine, namely thymine (T) or cytosine (C). Since the four different nucleotides differ only in their base, the abbreviations A, G, T and C are also used for the corresponding nucleotides.
The five carbon atoms of a deoxyribose are numbered from 1′ to 5′. The base is bound to the 1′ end of this sugar. The phosphate residue is attached to the 5′ end. Strictly speaking, deoxyribose is 2-deoxyribose; the name comes from the fact that, compared to a ribose molecule, an alcoholic hydroxy group (OH group) is missing at the 2′ position (i.e. has been replaced by a hydrogen atom).
At the 3′-position there is an OH group which links the deoxyribose to the 5′-carbon atom of the sugar of the next nucleotide via a so-called phosphodiester bond. Thus, each so-called single strand has two different ends: a 5′ and a 3′ end. DNA polymerases that synthesize DNA strands in the living world can only attach new nucleotides to the OH group at the 3′ end, but not at the 5′ end. The single strand therefore always grows from 5′ to 3′. A nucleoside triphosphate (with three phosphate residues) is supplied as a new building block, from which two phosphates are split off in the form of pyrophosphate. The remaining phosphate residue of each newly added nucleotide is connected to the OH group at the 3′ end of the last nucleotide present in the strand by splitting off water. The sequence of bases in the strand encodes the genetic information.
The double helix
DNA normally occurs as a helical double helix in a conformation called B-DNA. Two of the single strands described above are attached to each other in opposite directions: at each end of the double helix, one of the two single strands has its 3′ end, the other its 5′ end. Because of the juxtaposition, two specific bases are always opposite each other in the middle of the double helix, they are “paired”. The double helix is mainly stabilized by stacking interactions between successive bases of the same strand (and not, as often claimed, by hydrogen bonds between the strands).
Adenine and thymine are always paired, forming two hydrogen bonds, or cytosine with guanine, which are connected by three hydrogen bonds. A bridge is formed between the molecular positions 1═1 and 6═6, and in the case of guanine-cytosine pairings additionally between 2═2. Since the same bases are always paired, the sequence of the bases in one strand can be used to derive those of the other strand, the sequences are complementary. The hydrogen bonds are almost exclusively responsible for the specificity of the pairing, but not for the stability of the double helix.
Since a purine is always combined with a pyrimidine, the distance between the strands is the same everywhere, a regular structure is formed. The whole helix has a diameter of about 2 nm and winds further by 0.34 nm with each sugar molecule.
The planes of the sugar molecules are at an angle of 36° to each other, and full rotation is therefore achieved after 10 bases (360°) and 3.4 nm. DNA molecules can become very large. For example, the largest human chromosome contains 247 million base pairs.
When the two individual strands are twisted together, lateral gaps remain, so that the bases here are directly on the surface. There are two of these grooves that wind around the double helix (see illustrations and animation at the beginning of the article). The “large furrow” is 2.2 nm wide, the “small furrow” only 1.2 nm.
Accordingly, the bases in the large furrow are more accessible. Proteins that bind to the DNA in a sequence-specific manner, such as transcription factors, therefore usually bind to the large groove.
Some DNA dyes, for example DAPI, also bind to a furrow.
The cumulative binding energy between the two single strands holds them together. Covalent bonds are not present here, which means that the DNA double helix does not consist of one molecule, but two. This allows the two strands to be temporarily separated in biological processes.
In addition to the B-DNA just described, there is also A-DNA and a left-handed, so-called Z-DNA, which was first studied in 1979 by Alexander Rich and his colleagues at MIT. This occurs particularly in G-C-rich sections. It was not until 2005 that a crystal structure was reported which shows Z-DNA directly in a compound with B-DNA and thus provides evidence for the biological activity of Z-DNA. The following table and the adjacent figure show the differences between the three forms in direct comparison.
The base stackings are not exactly parallel to each other like books, but form wedges that tilt the helix in one direction or the other. The largest wedge is formed by adenosines paired with thymidines of the other strand. Consequently, a series of AT pairs forms an arc in the helix. When such series follow each other at short intervals, the DNA molecule assumes a curved or bent structure, which is stable. This is also called sequence-induced diffraction, since diffraction can also be induced by proteins (so-called protein-induced diffraction). Sequence-induced diffraction is often found at important locations in the genome.
Chromatin and chromosomes
Human chromosomes in the late metaphase of mitotic cell nucleus division: Each chromosome shows two chromatids, which are separated in the anaphase and divided for two cell nuclei.
In the eukaryotic cell, DNA is organised in the form of chromatin threads, called chromosomes, which are located in the cell nucleus. A single chromosome contains a long, continuous DNA double strand (in a chromatid) from the anaphase to the beginning of the S phase. At the end of the S phase, the chromosome consists of two identical DNA strands (in two chromatids).
Since such a DNA thread can be several centimetres long, but a cell nucleus is only a few micrometres in diameter, the DNA must be additionally compressed or “packed”. In eukaryotes, this is done with so-called chromatin proteins, of which the basic histones are particularly noteworthy. They form the nucleosomes around which the DNA is wrapped at the lowest packaging level. During nuclear division (mitosis), each chromosome is condensed to its maximum compact form. This allows them to be identified particularly well under the light microscope during the metaphase.
Bacterial and viral DNA
In prokaryotic cells, the majority of double-stranded DNA in the cases documented so far is not present as linear strands with a beginning and an end each, but as circular molecules – each molecule (i.e. each DNA strand) closes a circle with its 3′ and 5′ end. These two circular, closed DNA molecules are called bacterial chromosome or plasmid, depending on the length of the sequence. In bacteria, they are also not located in a cell nucleus, but are freely present in the plasma. The prokaryotic DNA is wound up into simple “supercoils” with the help of enzymes (for example topoisomerases and gyrases), which resemble a ringed telephone cord. By still rotating the helices around themselves, the space required for the genetic information is reduced. In the bacteria, topoisomerases ensure that the twisted double strand is wrested at a desired location by constantly cutting and rejoining the DNA (prerequisite for DNA transcription and DNA replication). Depending on the type, viruses contain either DNA or RNA as genetic information. In both DNA and RNA viruses, the nucleic acid is protected by a protein envelope.
Chemical and physical properties of the DNA double helix structure
At neutral pH, DNA is a negatively charged molecule, with the negative charges being located on the phosphate groups in the backbone of the strands. Although two of the three acidic OH groups of the phosphates are esterified with the respective neighbouring deoxyriboses, the third is still present and releases a proton at neutral pH, which causes the negative charge. This property is used in agarose gel electrophoresis to separate different DNA strands according to their length. Some physical properties such as the free energy and the melting point of the DNA are directly related to the GC content, i.e. they are sequence-dependent.
DNA Stack interactions
Two main factors are responsible for the stability of the double helix: base pairing between complementary bases and stacking interactions between consecutive bases.
Contrary to the initial assumption, the energy gain through hydrogen bonding is negligible, since the bases can form similarly good hydrogen bonds with the surrounding water. The hydrogen bonds of a GC base pair contribute only minimally to the stability of the double helix, while those of an AT base pair even have a destabilizing effect. Stack interactions, on the other hand, only have an effect in the double helix between successive base pairs: Between the aromatic ring systems of the heterocyclic bases, a dipole-induced dipole interaction is formed, which is energetically favorable. Thus, the formation of the first base pair is quite unfavorable due to the low energy gain and loss, but the elongation (lengthening) of the helix is energetically favorable, since the base pair stacking takes place under energy gain.
However, the stacking interactions are sequence-dependent and energetically most favorable for stacked GC-GC, less favorable for stacked AT-AT. The differences in the stacking interactions mainly explain why GC-rich DNA sections are thermodynamically more stable than AT-rich sections, while hydrogen bonding plays a minor role.
DNA melting point
The melting point of DNA is the temperature at which the binding forces between the two single strands are overcome and they separate from each other. This is also called denaturation.
As long as the DNA is denatured in a cooperative transition (which takes place in a narrow temperature range), the melting point is the temperature at which half of the double strands are denatured into single strands. From this definition the correct terms “midpoint of transition temperature” Tm are derived.
The melting point depends on the respective base sequence in the helix. It increases if there are more GC base pairs in the helix, since these are entropically more favourable than AT base pairs. This is not so much due to the different number of hydrogen bonds formed by the two pairs, but rather to the different stacking interactions. The stacking energy of two base pairs is much smaller if one of the two pairs is an AT base pair. GC stacks, on the other hand, are energetically more favorable and stabilize the double helix more strongly. The ratio of the GC base pairs to the total number of all base pairs is given by the GC content.
Since single-stranded DNA absorbs UV light about 40 percent more strongly than double-stranded DNA, the transition temperature can be easily determined in a photometer.
If the temperature of the solution falls below Tm, the single strands can rejoin each other. This process is called renaturation or hybridization. The interplay of de- and renaturation is exploited in many biotechnological processes, for example in polymerase chain reaction (PCR), Southern blots and in situ hybridisation.
Did you like this blog post? You can find more post about DNA, genetics and sequencing here!
You might be interested in the following related blog posts: