Table of Contents
What is Whole Exome Sequencing?
Whole exome sequencing (WES) is the sequencing of the exome, all protein-coding genes in the genome. This process involves two steps. The first step is to target sequences in the human exome only. In human genetics, these target regions are about 60 million base pairs, or about 1% of the human reference genome. The second step is to sequence the DNA using any high-throughput DNA sequencing platform and analyze the results.
Exome sequencing makes it possible to detect genetic changes that lead to changes in protein sequences, which can in turn lead to diseases such as atherosclerosis, Alzheimer’s disease, and others (you can read more about Alzheimer’s disease and dementia in our Research Library). Many diseases occur in the protein-coding region of the genome and this process allows users to learn more about their predisposition to disease than single gene analysis. However, it is now known that DNA variations outside the exons can affect gene activity and protein production and also lead to genetic disorders. Whole exome sequencing would miss these variations.
In the past, the main advantage of exome sequencing was the ability to perform greater gene screening and detect disease-associated mutations, such as copy number variants, in a way that was cost effective and consumed less time than whole-genome sequencing. However, thanks to advances in sequencing technology, whole genome sequencing is now just as fast and just as affordable. Nebula Genomics offers the most affordable 30X whole genome sequencing at $299 or less. We decode 100% of your DNA, not just 1% as with whole exome sequencing, providing you the most comprehensive view of your DNA.
Performing Whole Exome Sequencing
Whole exome sequencing steps start with DNA isolation and target enrichment and ends with DNA sequencing and analysis.
DNA isolation and enrichment strategies
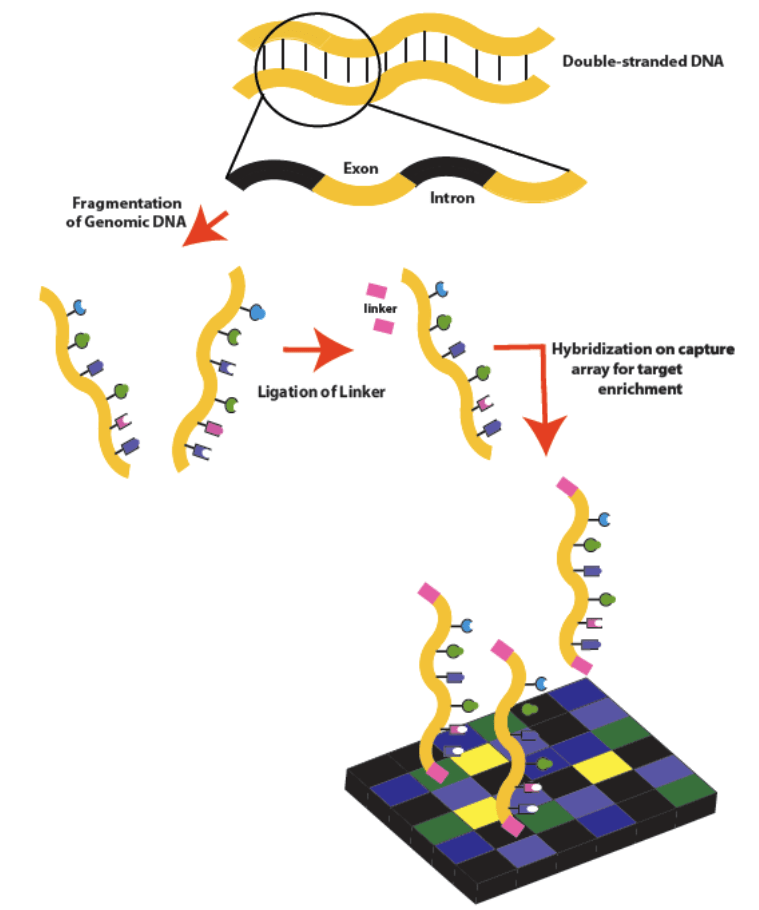
Whole exome sequencing labs isolate DNA from a provided sample, such as a saliva sample or cheek swab. The most common form of DNA isolation is phenol-chloroform extraction.
There are several strategies to enrich DNA from samples. The method used is often dependent on the sample and the desired result.
- PCR: Polymerase Chain Reaction (PCR) is the most common way to amplify desired DNA fragments for sequencing.
- Molecular inversion: This method involves closing the region of interest in a ring. A single-stranded DNA oligonucleotide primer contains a central part with a universal sequence with restriction sites, while the ends are complementary to the two sections of genomic DNA between which the sequence of interest is located.
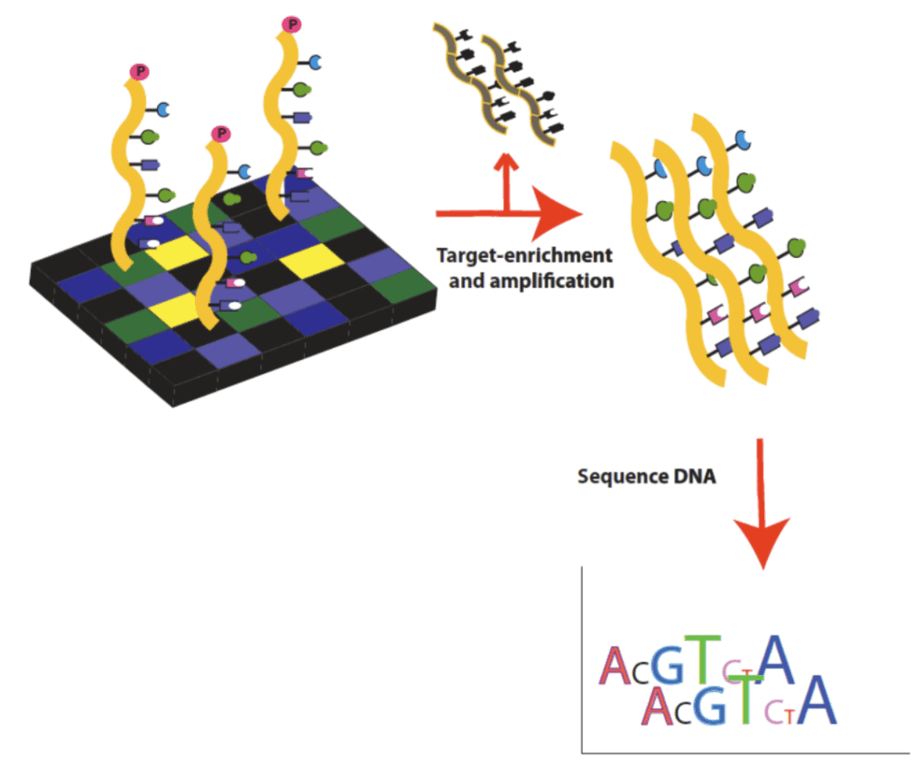
- Hybridization, array-based capture: microarrays are used that contain single-stranded oligonucleotides attached to a substrate with sequences from the genome capable of covering the sites of interest. Genomic DNA is cut into fragments, the ends of the fragments are made blunt, and universal primers are added. After hybridization of the fragments with probes on microarrays, the unhybridized fragments are washed from the substrate, and the remaining fragments are then amplified using PCR
- In-solution capture: A set of probes is synthesized in solution and fixed on streptavidin beads. The beads are placed in a solution with fragmented genomic DNA, where the probes are selectively hybridized to the desired genomic sites, after which the beads with the fragments of interest are precipitated and washed. The sections are then sequenced.
DNA Sequencing
There are many Next Generation Sequencing (NGS) technology platforms available. Some include the Roche 454 sequencer and Life Technologies SOLiD systems, the Life Technologies Ion Torrent and Illumina’s Illumina Genome Analyzer II (defunct) and subsequent Illumina MiSeq, HiSeq, and NovaSeq series instruments. ‘Short read’ NGS systems are ideal in analyzing many relatively short stretches of DNA sequence, as found in the exome.
Data Analysis
Even decoding just 1% of the human genome generates whole exome sequencing data sizes of around 8 GB and thus requires heavy data analysis, for which there are numerous programs to align and assemble reads. Some strategies that are used to improve the quality of data generation include:
- Comparing the genetic variants identified between sequencing and array-based genotyping (if possible)
- Comparing the coding SNPs to a whole genome sequenced individual with the disorder
- Comparing the coding SNPs with Sanger sequencing of HapMap individuals
To create variant calls, analysis programs identify variants from sequence data to create various files. Some of the common files are FASTQ, BAM/CRAM, and VCF files.
You can read more about the whole exome sequencing procedure on the whole exome sequencing wiki page.
Who should get tested?
The American College of Medical Genetics and Genomics has policies regarding when sequencing methods should be used, what results may arise, and what the results might indicate.
Specifically, they recommend that genetic testing be considered for certain individuals including:
- The phenotype or family history data strongly implicate a genetic etiology, but the phenotype does not correspond with a specific disorder for which a genetic test targeting a specific gene is available on a clinical basis
- A patient presents with a defined genetic disorder that demonstrates a high degree of genetic heterogeneity, making WES or WGS analysis of multiple genes simultaneously a more practical approach
- A patient presents with a likely genetic disorder but specific genetic tests available for that phenotype have failed to arrive at a diagnosis
- A fetus with a likely genetic disorder in which specific genetic tests
Advantages of Whole Exome Sequencing
Whole exome sequencing technology, like other DNA testing methods, aims to use the genome to help diagnose and guide treatments for mendelian disorders. It also provides users information that can help them advise family members of their risk.
This method has several advantages over single gene analysis, one of the most common methods of DNA sequencing. First, whole exome sequencing provides the ability to identify a wide range of mutations in candidate genes that are not tested. In this way, a user can identify disease genes they may not have suspected they were at risk for. Secondly, in the clinic, whole exome sequencing provides the ability to identify cases where mutations from different genes contribute to the different phenotypes in the same patient.
Whole exome sequencing can also be used in scientific research to identify inherited rare diseases not detectable by microarrays. For example, the allele variants for several severe disorders are known to be located on the exons but are also extremely rare in the population. Thus, standard genotyping assays that need large allele databases to be effective often miss these variants. Multiple case studies highlight the usefulness of the whole exome sequencing approach. In 2009, a group of scientists used whole exome sequencing to identify the mutation that causes Bartter syndrome and congenital chloride diarrhea.
In 2011, Ambry Genetics was the first company to offer an exome sequencing service for clinical disease diagnostics. The company claims that exome sequencing results will allow them to diagnose diseases for which traditional diagnostic approaches are not applicable. You can read more about the history and results of Ambry Genetics in our product review!

Limitations of Whole Exome Sequencing
Whole exome sequencing and whole genome sequencing: Both of these procedures are next generation sequencing technologies. While whole exome sequencing demonstrates some advantages over microarray-based genotyping for clinical analysis of disease risk, it also has limitations. The most important difference between whole exome and whole genome sequencing is the amount of DNA sequenced. Since whole exome sequencing decodes only 1% of the genome, it fails to analyze the other 99%, including structural and non-coding regions. On the other hand, Whole Genome Sequencing reveals the full 100% of your DNA that includes all 3 billion base pairs (only 40 million base pairs are covered with whole exome sequencing).
How much does whole exome sequencing cost? For a while, whole exome sequencing was considered the most beneficial type of DNA sequencing as it provided an expanded view of DNA coding regions compared to microarray genotyping at a much lower price than whole genome sequencing. However, this is no longer the case.
Whole genome sequencing can be achieved for a fraction of its original price and provide more information than whole exome sequencing. At Nebula Genomics, you can get your entire genome sequenced for less than $299. This is comparable and even cheaper than many at-home whole exome sequencing products.
Chromosomal Microarray vs Whole Exome Sequencing vs Whole Genome Sequencing
| Microarray Genotyping | Whole exome sequencing | Whole genome sequencing | |
| Purpose | Identify specific variants | Decode all genes | Decode all genes as well as all structural and non-coding regions |
| Throughput | Low-throughput | High-throughput | High-throughput |
| Requires prior identification of variant | Yes | No | No |
| Amount of coverage | 0.5 million positions (0.02% of the genome) | 60 million positions (~ 1% of the genome) | 6 billion positions (~ 100% of the genome) |
| Cost | ~ $100 | ~ $300 | ~ $1000 (but Nebula Genomics offers it for $299) |
Direct to consumer whole exome sequencing companies
Various companies sell whole exome sequencing products. Some of these products must be ordered through a healthcare provider and may be covered by insurance. These tests are often used to diagnose disease. Some companies offer the service as direct to consumer at-home tests which are analyzed in CLIA approved and CAP accredited labs so while they can’t be used as diagnostic tools, they can be brought to healthcare providers for consultation.
23andMe ran a pilot whole exome sequencing program that was announced in September 2011 and was discontinued in 2012. Consumers could obtain data at a whole exome sequencing cost of $999. The company provided raw data, and did not offer analysis.
- Ambry genetics was the first company to offer an exome sequencing service for clinical disease diagnostics. Today, customers can order tests through healthcare providers and most of the cost is covered by insurance.
- CircleDNA offers at home WES for $189 – $629 and many tests include genetic counseling. This company also collaborates with DNAFit.
- Dante Labs’ WES starts at $849, at a much higher price than competitors. This company also offers whole genome sequencing starting at $599.
- Full Genomes offers WES starting at $525.
- GeneDx began offering WES in 2012. Their prices are variable.
- HelixDNA offers WES at a reasonable price of $145 plus an additional $299 for a useful app.
- Both Invitae WES and Myriad Genetics offer diagnostic tests. These tests, while expensive, may be covered by insurance.
- Tellmegen looks at select regions of the exome known to be linked to disease and health. Tests cost $235.
You can read many more company reviews on our blog and check out our complete guide to the best DNA test kit and other home tests.
